# Initialize Model
model_seirconn <- ModelSEIRCONN(
name = "Flu",
n = 10000,
prevalence = 0.01,
contact_rate = 2,
transmission_rate = 0.5,
incubation_days = 2,
recovery_rate = 1/7
)
# Creating a tool
epitool <- tool(
name = "Vaccine",
susceptibility_reduction = .9,
transmission_reduction = .5,
recovery_enhancer = .5,
death_reduction = .9
)
# Adding a global action
vaccine_day_10 <- globalaction_tool(epitool, prob = .5, day = 10)
add_global_action(model_seirconn, vaccine_day_10)
# Generating a saver
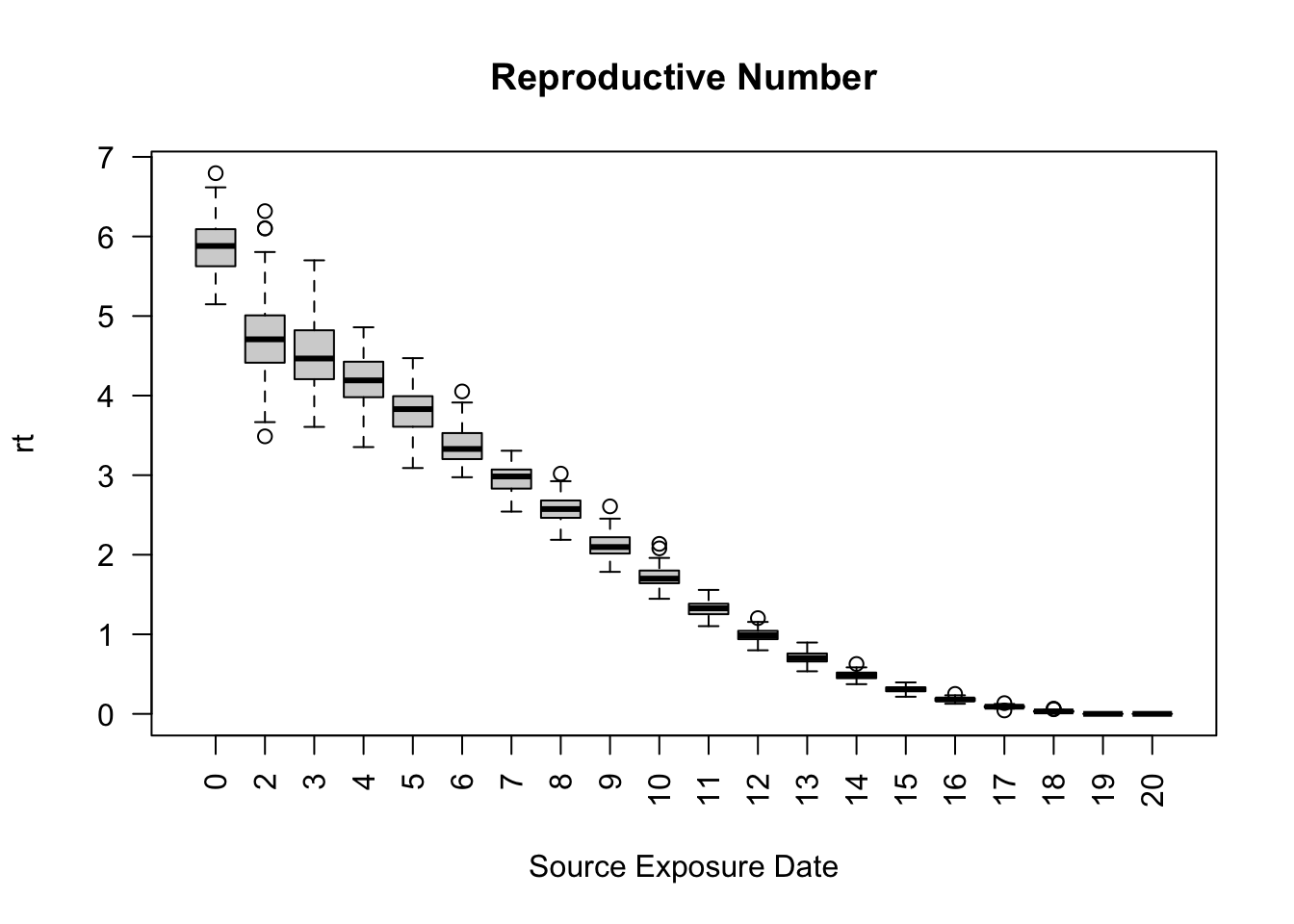
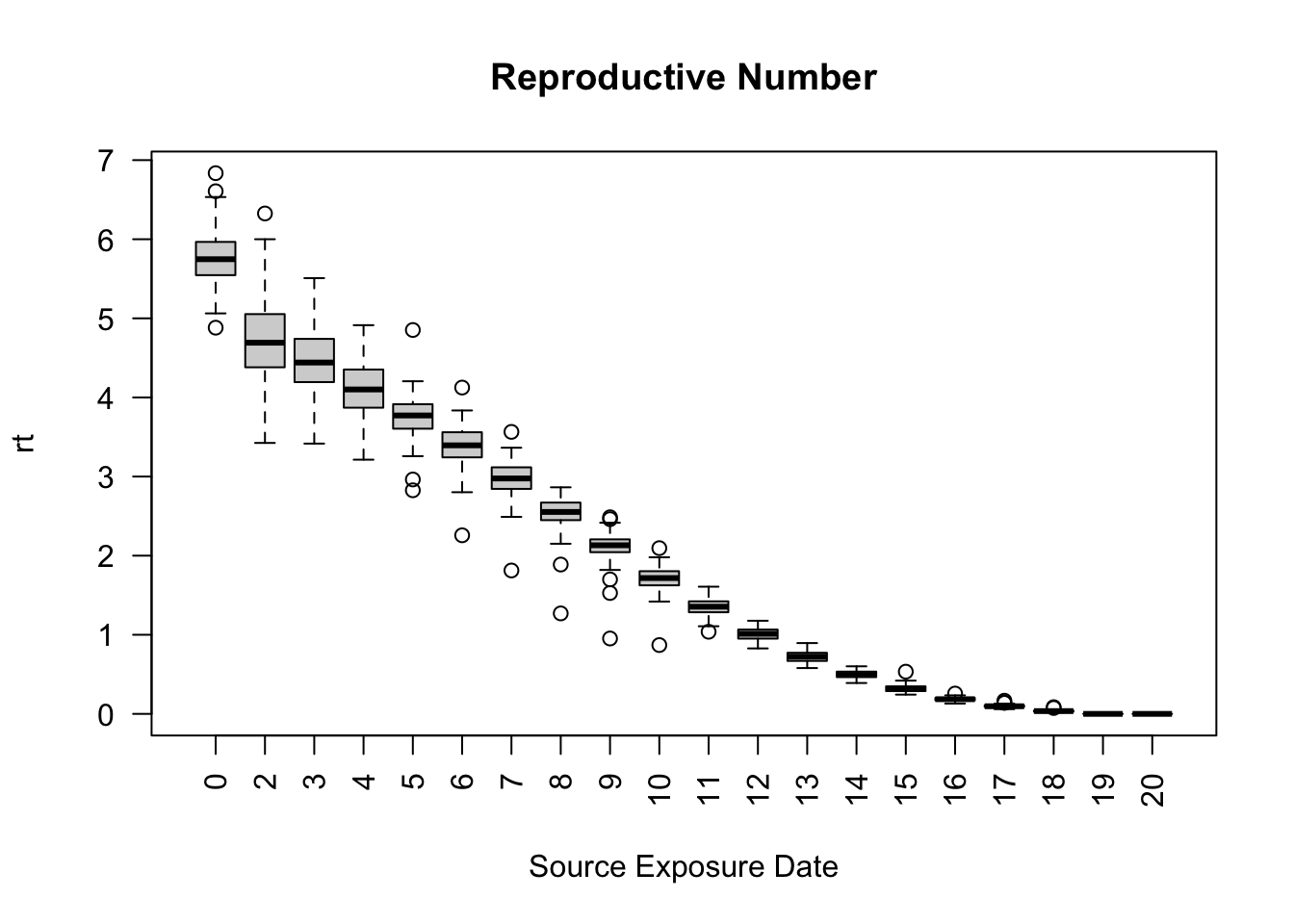
saver <- make_saver("reproductive")
# Running the model
run_multiple(model_seirconn, ndays = 20, nsims = 100, saver = saver,
nthread = 2)