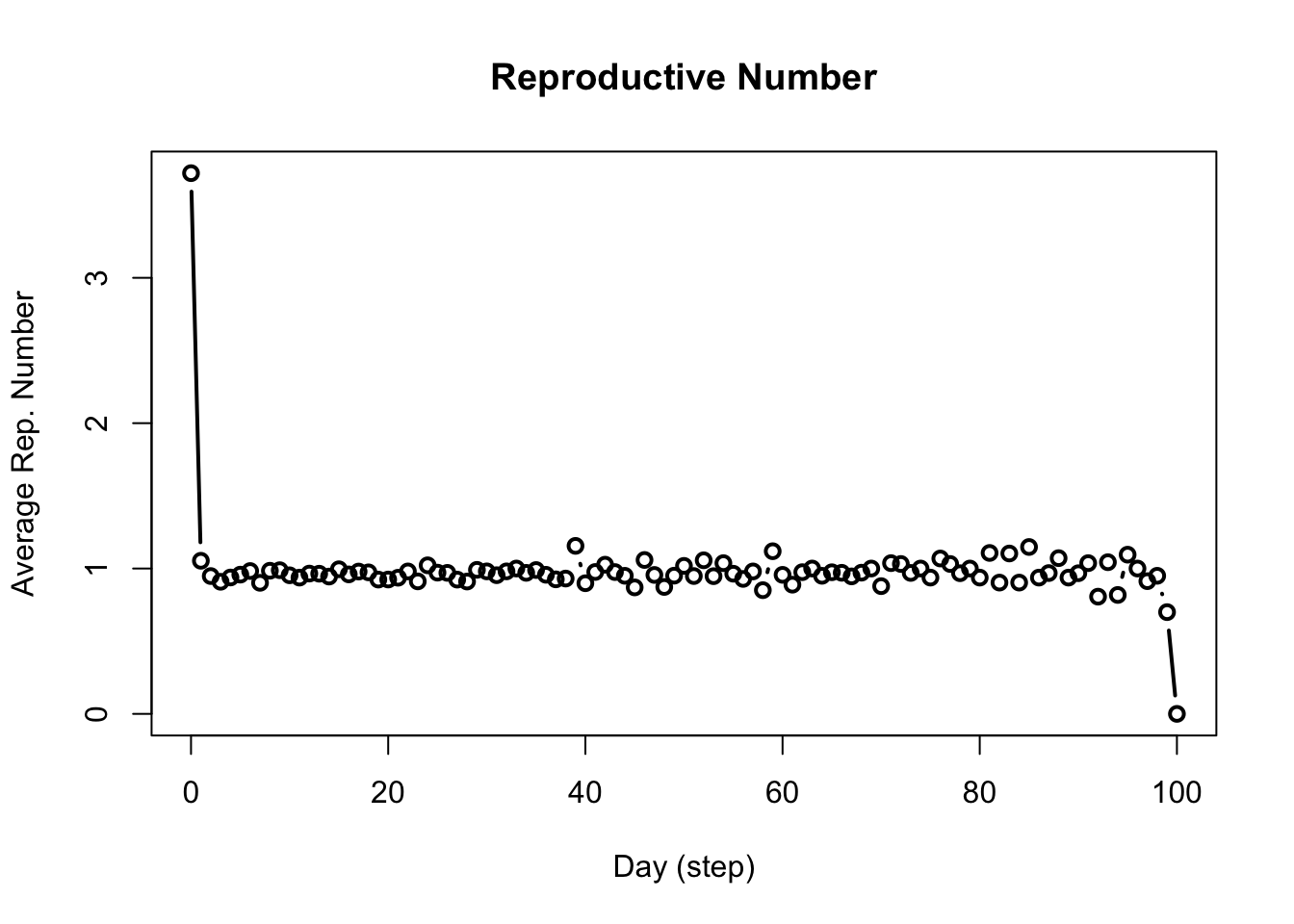
library(epiworldR)
# Initialize model
model_seir <- ModelSEIR("COVID-19",
prevalence = 0.01,
transmission_rate = 0.9,
recovery_rate = 1/4,
incubation_days = 4)
# Adding a small world population
agents_smallworld(
model_seir,
n = 10000,
k = 5,
d = FALSE,
p = .01
)
# Running model
run(model_seir, ndays = 100, seed = 1912)_________________________________________________________________________
Running the model...
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
done.# Plotting
plot(model_seir)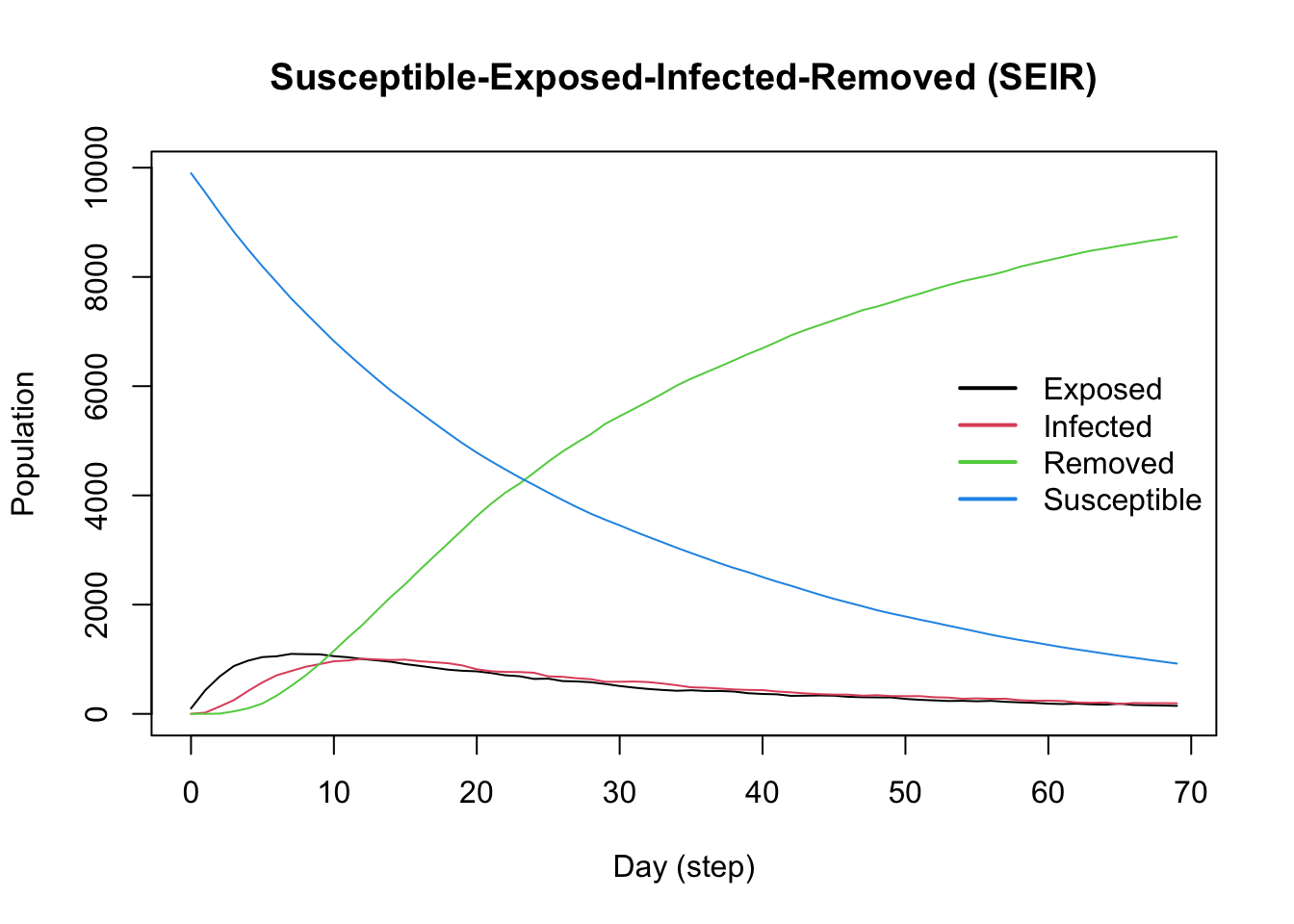
# Day of peak infections
x <- get_hist_total(model_seir)
which.max(x$counts[x$state == "Infected"]) - 1[1] 12# Number of infections on peak day
max(x$counts[x$state == "Infected"])[1] 1010